Publications
For a full list see Google Scholar
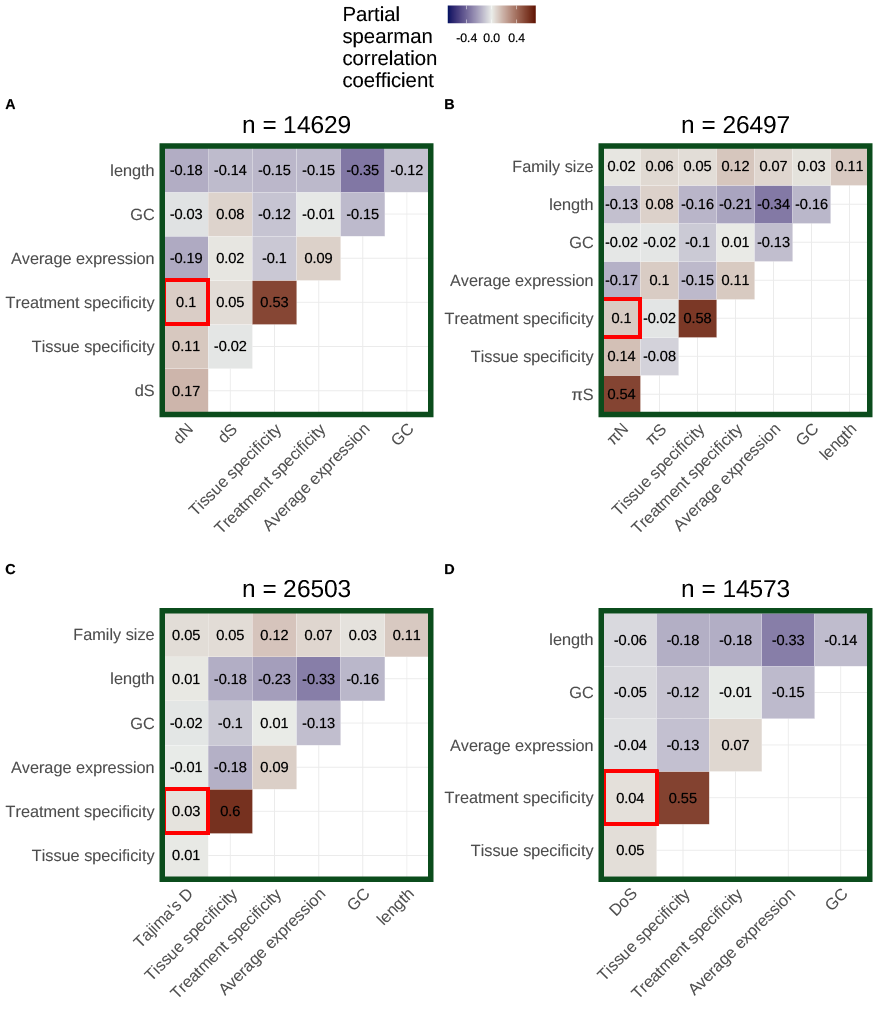
Miles Roberts, EB Josephs
Genetics (2023)
Using 27 terabases of publicly available RNA-sequencing data, we quantified the environment-specificity of gene expression for every gene in the Arabidopsis thaliana genome. We found that genes with more environment-specific expression are under weaker negative selection on average, highlighting potential limits to the evolution of expression plasticity.
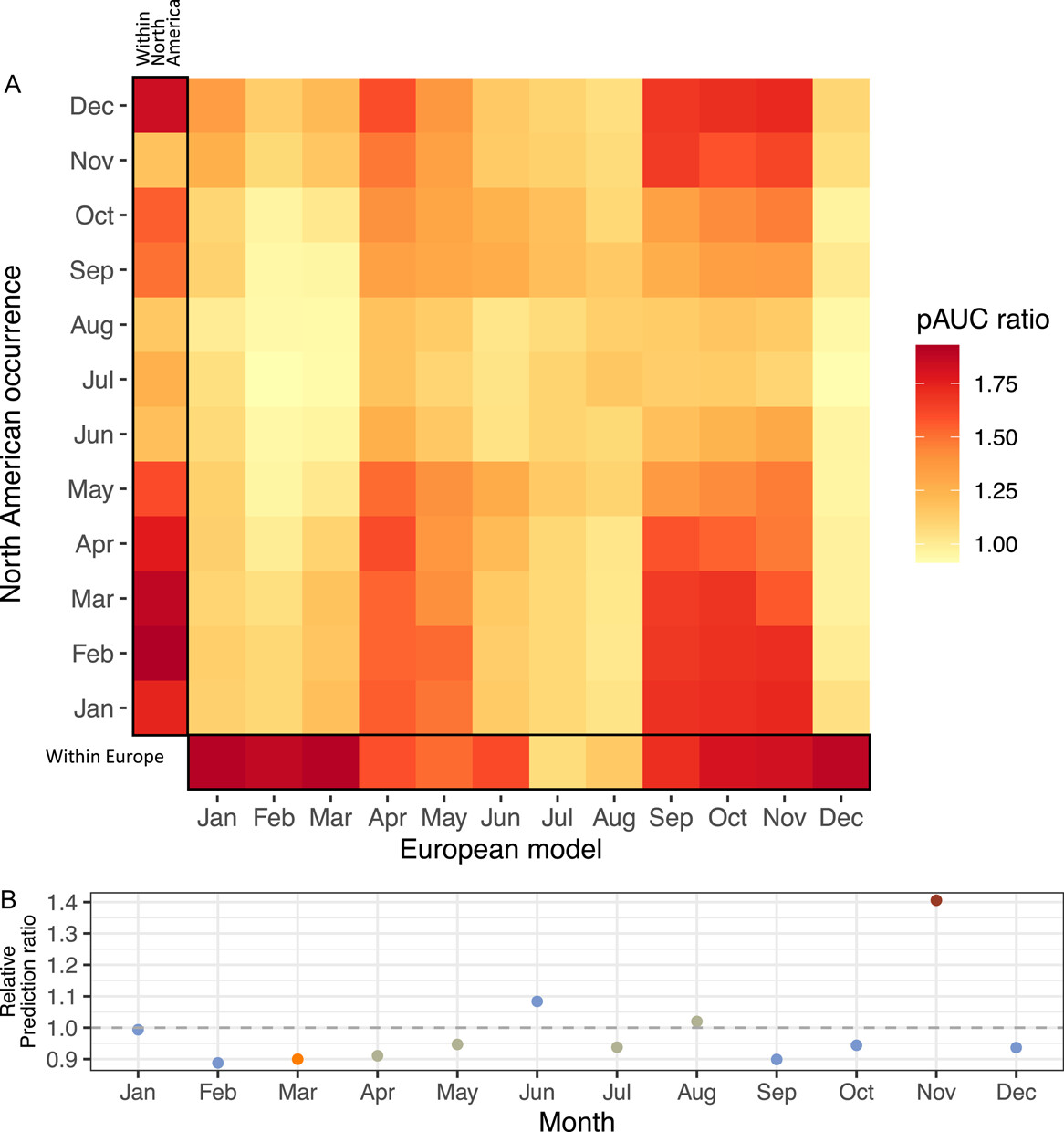
MK Wilson Brown, EB Josephs
AJB (2023)
We generated full-year and monthy niche models for Capsella in Europe and North America. We find full-year ecological niche models have low transferability across continents and there are continental differences in the climate conditions that influence the distribution of C. bursa-pastoris. Monthly models have greater predictive accuracy than full-year models in cooler seasons, but no monthly models can predict North American summer occurrences very well.

EB Josephs, ML Van Etten, A Harkess, A Platts, RS Baucom
Evolution Letters (2021)
In collaboration with Gina Baucom’s lab at the University of Michigan, we investigated the potential for gene expression responses to herbicide to be adaptive. We used gene expression data from plants that had been grown with and without herbicide, along with expression data from artificial selection experiments for herbicide resistance to show that expression responses are often likely beneficial
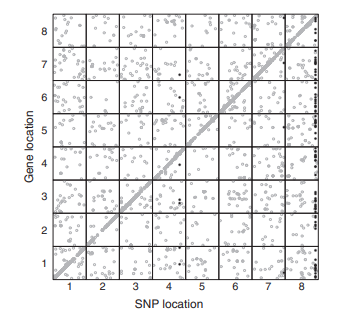
EB Josephs, YW Lee, CW Wood, DJ Schoen, SI Wright, and JR Stinchcombe
MBE (2020)
We detected loci that affect the expression of nearby genes (cis-eQTLs) and of far away genes (trans-eQTLs) and compared their allele frequencies and other signatures of selection to show that negative selection acts more strongly on trans-eQTLs than cis-eQTLs. We also developed a method using coexpression networks to identify loci that affected the expression of large modules of genes with correlated expression
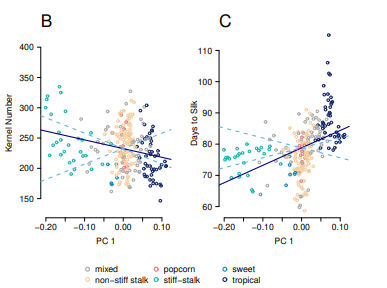
EB Josephs, JJ Berg, J Ross-Ibarra, G Coop
Genetics (2019)
We developed a method to detect local adaptation in diverse samples that have been genotyped and phenotyped in common garden. Specifically, we look for excess trait differentiation along the principal components of relatedness beyond an expectation based on neutral genetic drift. We then applied these methods to detect adaptation in domesticated maize.
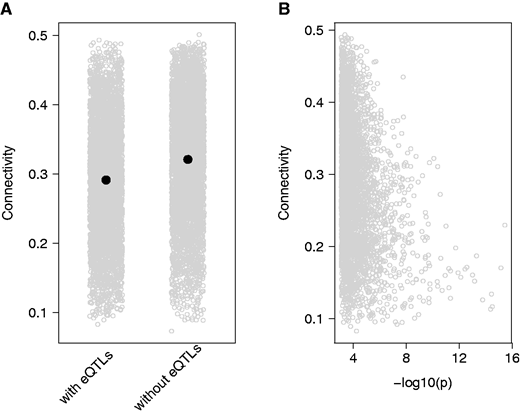
EB Josephs, SI Wright, JR Stinchcombe, DJ Schoen
Genome Biology and Evolution (2017)
We used leaf expression data to construct coexpression networks that describe the correlation in expression between genes. Connectivity in these networks was associated with both the strength of negative selection on gene sequence and the presence of local regulatory variants (‘eQTLs’)
EB Josephs, JR Stinchcombe, SI Wright
New Phytologist (2017)
A review of techniques and best practices for understanding the maintenance of genetic variation using population and quantitative genetics
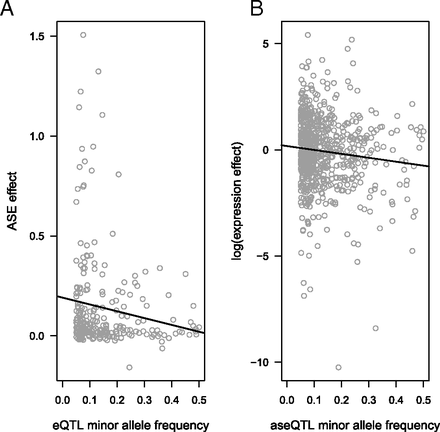
EB Josephs, YW Lee, JR Stinchcombe, SI Wright
PNAS (2015)
We identified genetic variants that control gene expression and showed that purifying (or negative) selection acts on these variants based on patterns of allele frequency and effect sizes.
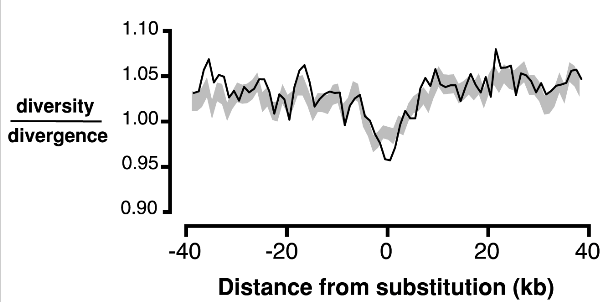
RJ Williamson, EB Josephs, AE Platts, KM Hazzouri, A Haudry, M Blanchette, SI Wright.
Plos Genetics (2014)
We analyzed population genetic signatures of selection in Capsella and showed that selection is important in shaping genomic variation.